Publications
Group highlights
At the end of this page, you can find the full list of publications. See also BRL@UCY papers in PubMed.
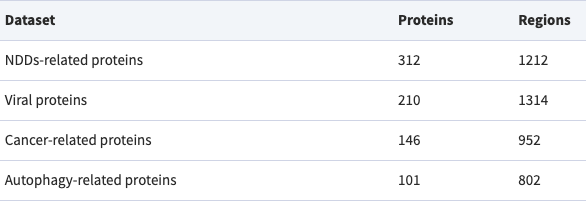
This paper presents the latest version of the DisProt database, the gold standard database for intrinsically disordered proteins and regions, also introducing the thematic dataset on Autophagy proteins, with the contribution of BRL members.
Aspromonte MC, Nugnes MV, Quaglia F, Bouharoua A; DisProt Consortium; Tosatto SCE, Piovesan D.
Nucleic Acids Res. 2024;52(D1):D434-D441.
Resource available at https://disprot.org. Direct access to the Autophagy protein thematic dataset.
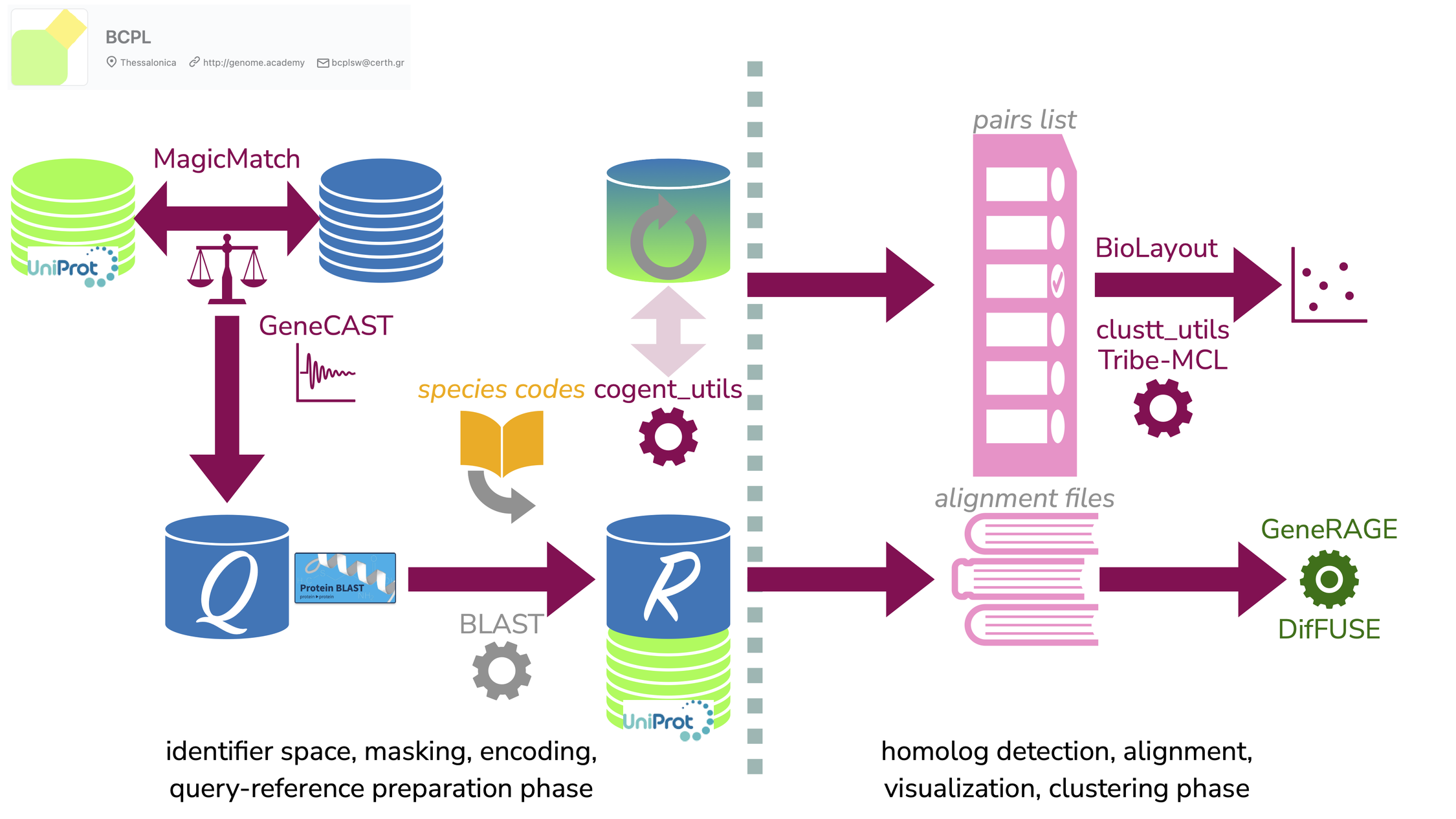
This paper describes the re-launch of a range of established software components for computational genomics, made available online as open source.
Vasileiou D, Karapiperis C, Baltsavia I, Chasapi A, Ahrén D, Janssen PJ, Iliopoulos I, Promponas VJ, Enright AJ, Ouzounis CA.
PLoS Comput Biol. 2023 Nov 7;19(11):e1011498.
Resource available at https://github.com/bcpl-certh/cgg-toolkit.

Several selective macroautophagy receptor and adaptor proteins bind members of the Atg8 (autophagy related 8) family using short linear motifs (SLiMs), most often referred to as Atg8-family interacting motifs (AIMs) or LC3-interacting regions (LIRs). AIM/LIR motifs have been extensively studied during the last fifteen years, since they can uncover the underlying biological mechanisms and possible substrates for this key catabolic process of eukaryotic cells. Prompted by the fact that experimental information regarding LIR motifs can be found scattered across heterogeneous literature resources, we have developed LIRcentral, a freely available online repository for user-friendly access to comprehensive, high-quality information regarding LIR motifs from manually curated publications.
Chatzichristofi A, Sagris V, Pallaris A, Eftychiou M, Kalvari I, Price N, Theodosiou T, Iliopoulos I, Nezis IP, Promponas VJ.
Resource available at https://lircentral.eu.
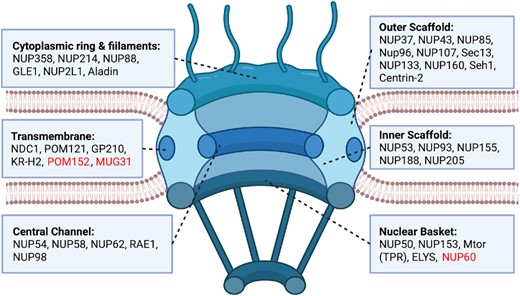
Nuclear Pore Complex subunits (nucleoporins or ‘NUPs’) are often difficult to characterize based on sequence similarity. Here, we describe a robust library of nucleoporin protein sequences and their respective family-specific position-specific scoring matrices which can be used to effectively characterize NUPs across eukaryotic sequence datasets.
Ioannides AN, Katsani KR, Ouzounis CA, Promponas VJ.
NAR Genom Bioinform. 2023;5(1):lqad025.
Resource available at https://doi.org/10.5281/zenodo.7684166.
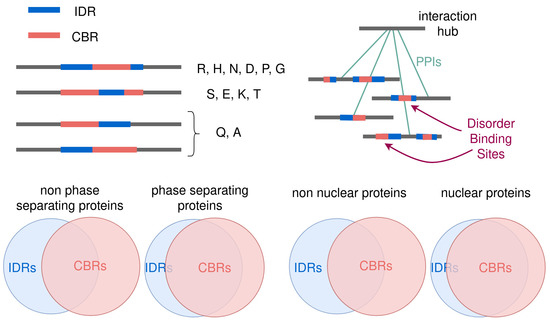
Intrinsically disordered regions (IDRs) in protein sequences are flexible, have low structural constraints and as a result have faster rates of evolution. This lack of evolutionary conservation greatly limits the use of sequence homology for the classification and functional assessment of IDRs, as opposed to globular domains. The study of IDRs requires other properties for their classification and functional prediction. While composition bias is not a necessary property of IDRs, compositionally biased regions (CBRs) have been noted as frequent part of IDRs. We hypothesized that to characterize IDRs, it could be helpful to study their overlap with particular types of CBRs. Here, we evaluate this overlap in the human proteome and discovering interesting trends, opening ways to pin down the function of IDRs from their partial compositional biases.
Kastano K, Mier P, Dosztányi Z, Promponas VJ, Andrade-Navarro MA.
Biomolecules. 2022;12(10):1486.
Full List of publications
A paradigm for post-embryonic Oct4 re-expression: E7-induced hydroxymethylation regulates Oct4 expression in cervical cancer.
Panayiotou T, Eftychiou M, Patera E, Promponas VJ, Strati K.
J Med Virol. 2023 Dec;95(12):e29264.
DisProt in 2024: improving function annotation of intrinsically disordered proteins.
Aspromonte MC, Nugnes MV, Quaglia F, Bouharoua A; DisProt Consortium; Tosatto SCE, Piovesan D.
Nucleic Acids Res. 2024;52(D1):D434-D441.
A choice, not an obligation : Releasing scientific software as open source should be the responsibility of the authors.
Kappas I, Promponas VJ, Ouzounis CA.
EMBO Rep. 2024 Jan 2.
CGG toolkit: Software components for computational genomics.
Vasileiou D, Karapiperis C, Baltsavia I, Chasapi A, Ahrén D, Janssen PJ, Iliopoulos I, Promponas VJ, Enright AJ, Ouzounis CA.
PLoS Comput Biol. 2023 Nov 7;19(11):e1011498.
LIRcentral: a manually curated online database of experimentally validated functional LIR motifs.
Chatzichristofi A, Sagris V, Pallaris A, Eftychiou M, Kalvari I, Price N, Theodosiou T, Iliopoulos I, Nezis IP, Promponas VJ.
Autophagy. 2023:3189-3200.
A library of sensitive position-specific scoring matrices for high-throughput identification of nuclear pore complex subunits.
Ioannides AN, Katsani KR, Ouzounis CA, Promponas VJ.
NAR Genom Bioinform. 2023;5(1):lqad025.
Functional Tuning of Intrinsically Disordered Regions in Human Proteins by Composition Bias.
Kastano K, Mier P, Dosztányi Z, Promponas VJ, Andrade-Navarro MA.
Biomolecules. 2022;12(10):1486.
DisProt in 2022: improved quality and accessibility of protein intrinsic disorder annotation.
Quaglia F, Mészáros B, Salladini E, Hatos A, Pancsa R, Chemes LB, Pajkos M, Lazar T, Peña-Díaz S, Santos J, Ács V, Farahi N, Fichó E, Aspromonte MC, Bassot C, Chasapi A, Davey NE, Davidović R, Dobson L, Elofsson A, Erdős G, Gaudet P, Giglio M, Glavina J, Iserte J, Iglesias V, Kálmán Z, Lambrughi M, Leonardi E, Longhi S, Macedo-Ribeiro S, Maiani E, Marchetti J, Marino-Buslje C, Mészáros A, Monzon AM, Minervini G, Nadendla S, Nilsson JF, Novotný M, Ouzounis CA, Palopoli N, Papaleo E, Pereira PJB, Pozzati G, Promponas VJ, Pujols J, Rocha ACS, Salas M, Sawicki LR, Schad E, Shenoy A, Szaniszló T, Tsirigos KD, Veljkovic N, Parisi G, Ventura S, Dosztányi Z, Tompa P, Tosatto SCE, Piovesan D.
Nucleic Acids Res. 2022;50(D1):D480-D487
Solving the Protein Secondary Structure Prediction Problem With the Hessian Free Optimization Algorithm.
Charalampous K, Agathocleous M, Christodoulou C, Promponas VJ.
IEEE Access. 2022; 10:27759-27770.
Critical assessment of protein intrinsic disorder prediction.
Necci M, Piovesan D; CAID Predictors; DisProt Curators; Tosatto SCE.
Nat Methods. 2021;18(5):472-481.
Statistical and Machine Learning Techniques in Human Microbiome Studies: Contemporary Challenges and Solutions.
Moreno-Indias I, Lahti L, Nedyalkova M, Elbere I, Roshchupkin G, Adilovic M, Aydemir O, Bakir-Gungor B, Santa Pau EC, D’Elia D, Desai MS, Falquet L, Gundogdu A, Hron K, Klammsteiner T, Lopes MB, Marcos-Zambrano LJ, Marques C, Mason M, May P, Pašić L, Pio G, Pongor S, Promponas VJ, Przymus P, Saez-Rodriguez J, Sampri A, Shigdel R, Stres B, Suharoschi R, Truu J, Truică CO, Vilne B, Vlachakis D, Yilmaz E, Zeller G, Zomer AL, Gómez-Cabrero D, Claesson MJ.
Front Microbiol. 2021;12:635781.
Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition) 1
Klionsky DJ, Abdel-Aziz AK, Abdelfatah S, Abdellatif M, [..], Promponas VJ, [..], Stallings CL, Tong CK.
Autophagy. 2021;17(1):1-382.
Evolutionary Study of Disorder in Protein Sequences.
Kastano K, Erdős G, Mier P, Alanis-Lobato G, Promponas VJ, Dosztányi Z, Andrade-Navarro MA.
Biomolecules. 2020;10(10):1413.
Recognition of LD motifs by the focal adhesion targeting domains of focal adhesion kinase and proline-rich tyrosine kinase 2-beta: Insights from molecular dynamics simulations.
Michael E, Polydorides S, Promponas VJ, Skourides P, Archontis G.
Proteins. 2020.
PlaToLoCo: the first web meta-server for visualization and annotation of low complexity regions in proteins.
Jarnot P, Ziemska-Legiecka J, Dobson L, Merski M, Mier P, Andrade-Navarro MA, Hancock JM, Dosztányi Z, Paladin L, Necci M, Piovesan D, Tosatto SCE, Promponas VJ, Grynberg M, Gruca A.
Nucleic Acids Res. 2020;48(W1):W77-W84.
Pseudomonas aeruginosa core metabolism exerts a widespread growth-independent control on virulence.
Panayidou S, Georgiades K, Christofi T, Tamana S, Promponas VJ, Apidianakis Y.
Sci Rep. 2020;10(1):9505.
The bioinformatics wealth of nations.
Chasapi A, Promponas VJ, Ouzounis CA.
Bioinformatics. 2020;36(9):2963-2965.
Disentangling the complexity of low complexity proteins.
Mier P, Paladin L, Tamana S, Petrosian S, Hajdu-Soltész B, Urbanek A, Gruca A, Plewczynski D, Grynberg M, Bernadó P, Gáspári Z, Ouzounis CA, Promponas VJ, Kajava AV, Hancock JM, Tosatto SCE, Dosztanyi Z, Andrade-Navarro MA.
Brief Bioinform. 2020;21(2):458-472.
DisProt: intrinsic protein disorder annotation in 2020.
Hatos A, Hajdu-Soltész B, Monzon AM, Palopoli N, Álvarez L, Aykac-Fas B, Bassot C, Benítez GI, Bevilacqua M, Chasapi A, Chemes L, Davey NE, Davidović R, Dunker AK, Elofsson A, Gobeill J, Foutel NSG, Sudha G, Guharoy M, Horvath T, Iglesias V, Kajava AV, Kovacs OP, Lamb J, Lambrughi M, Lazar T, Leclercq JY, Leonardi E, Macedo-Ribeiro S, Macossay-Castillo M, Maiani E, Manso JA, Marino-Buslje C, Martínez-Pérez E, Mészáros B, Mičetić I, Minervini G, Murvai N, Necci M, Ouzounis CA, Pajkos M, Paladin L, Pancsa R, Papaleo E, Parisi G, Pasche E, Barbosa Pereira PJ, Promponas VJ, Pujols J, Quaglia F, Ruch P, Salvatore M, Schad E, Szabo B, Szaniszló T, Tamana S, Tantos A, Veljkovic N, Ventura S, Vranken W, Dosztányi Z, Tompa P, Tosatto SCE, Piovesan D.
Nucleic Acids Res. 202048(D1):D269-D276.
Establishment of computational biology in Greece and Cyprus: Past, present, and future.
Chasapi A, Aivaliotis M, Angelis L, Chanalaris A, Iliopoulos I, Kappas I, Karapiperis C, Kyrpides NC, Pafilis E, Panteris E, Topalis P, Tsiamis G, Vizirianakis IS, Vlassi M, Promponas VJ, Ouzounis CA.
PLoS Comput Biol. 2019;15(12):e1007532.
Tandem repeats lead to sequence assembly errors and impose multi-level challenges for genome and protein databases.
Tørresen OK, Star B, Mier P, Andrade-Navarro MA, Bateman A, Jarnot P, Gruca A, Grynberg M, Kajava AV, Promponas VJ, Anisimova M, Jakobsen KS, Linke D.
Nucleic Acids Res. 2019;47(21):10994-11006.
Low complexity regions in the proteins of prokaryotes perform important functional roles and are highly conserved.
Ntountoumi C, Vlastaridis P, Mossialos D, Stathopoulos C, Iliopoulos I, Promponas V, Oliver SG, Amoutzias GD.
Nucleic Acids Res. 2019;47(19):9998-10009.
An intrinsically disordered proteins community for ELIXIR.
Davey NE, Babu MM, Blackledge M, Bridge A, Capella-Gutierrez S, Dosztanyi Z, Drysdale R, Edwards RJ, Elofsson A, Felli IC, Gibson TJ, Gutmanas A, Hancock JM, Harrow J, Higgins D, Jeffries CM, Le Mercier P, Mészáros B, Necci M, Notredame C, Orchard S, Ouzounis CA, Pancsa R, Papaleo E, Pierattelli R, Piovesan D, Promponas VJ, Ruch P, Rustici G, Romero P, Sarntivijai S, Saunders G, Schuler B, Sharan M, Shields DC, Sussman JL, Tedds JA, Tompa P, Turewicz M, Vondrasek J, Vranken WF, Wallace BA, Wichapong K, Tosatto SCE.
F1000Res. 2019;8:ELIXIR-1753.
An updated view of the oligosaccharyltransferase complex in Plasmodium.
Tamana S, Promponas VJ.
Glycobiology. 2019;29(5):385-396.
Hypothesis, analysis and synthesis, it’s all Greek to me.
Iliopoulos I, Ananiadou S, Danchin A, Ioannidis JP, Katsikis PD, Ouzounis CA, Promponas VJ.
Elife. 2019;8:e43514.
A fragment of the alarmin prothymosin α as a novel biomarker in murine models of bacteria-induced sepsis.
Samara P, Miriagou V, Zachariadis M, Mavrofrydi O, Promponas VJ, Dedos SG, Papazafiri P, Kalbacher H, Voelter W, Tsitsilonis O.
Oncotarget. 2017 Jul 25;8(30):48635-48649.
Functional characterisation of long intergenic non-coding RNAs through genetic interaction profiling in Saccharomyces cerevisiae.
Kyriakou D, Stavrou E, Demosthenous P, Angelidou G, San Luis BJ, Boone C, Promponas VJ, Kirmizis A.
BMC Biol. 2016;14(1):106.
iLIR database: A web resource for LIR motif-containing proteins in eukaryotes.
Jacomin AC, Samavedam S, Promponas V, Nezis IP.
Autophagy. 2016;12(10):1945-1953.
Sequence evidence for common ancestry of eukaryotic endomembrane coatomers.
Promponas VJ, Katsani KR, Blencowe BJ, Ouzounis CA.
Sci Rep. 2016;6:22311.
Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition).
Klionsky DJ, Abdelmohsen K, Abe A, [..], Promponas VJ, [..], Zoladek T, Zong WX, Zorzano A, Zughaier SM.
Autophagy. 2016;12(1):1-222.
Annotation inconsistencies beyond sequence similarity-based function prediction - phylogeny and genome structure.
Promponas VJ, Iliopoulos I, Ouzounis CA.
Stand Genomic Sci. 2015;10:108.
Kirmitzoglou I, Promponas VJ. LCR-eXXXplorer: a web platform to search, visualize and share data for low complexity regions in protein sequences.
Bioinformatics. 2015;31(13):2208-10.
LiSIs: An Online Scientific Workflow System for Virtual Screening.
Kannas CC, Kalvari I, Lambrinidis G, Neophytou CM, Savva CG, Kirmitzoglou I, Antoniou Z, Achilleos KG, Scherf D, Pitta CA, Nicolaou CA, Mikros E, Promponas VJ, Gerhauser C, Mehta RG, Constantinou AI, Pattichis CS.
Comb Chem High Throughput Screen. 2015;18(3):281-95.
BioTextQuest(+): a knowledge integration platform for literature mining and concept discovery.
Papanikolaou N, Pavlopoulos GA, Pafilis E, Theodosiou T, Schneider R, Satagopam VP, Ouzounis CA, Eliopoulos AG, Promponas VJ, Iliopoulos I.
Bioinformatics. 2014;30(22):3249-56.
Guest editorial: Computational solutions to large-scale data management and analysis in translational and personalized medicine.
Tsiknakis M, Promponas VJ, Graf N, Wang MD, Wong ST, Bourbakis N, Pattichis CS.
IEEE J Biomed Health Inform. 2014;18(3):720-1.
iLIR: A web resource for prediction of Atg8-family interacting proteins.
Kalvari I, Tsompanis S, Mulakkal NC, Osgood R, Johansen T, Nezis IP, Promponas VJ.
Autophagy. 2014;10(5):913-25.
Experimental evidence validating the computational inference of functional associations from gene fusion events: a critical survey.
Promponas VJ, Ouzounis CA, Iliopoulos I.
Brief Bioinform. 2014;15(3):443-54.
Functional genomics evidence unearths new moonlighting roles of outer ring coat nucleoporins.
Katsani KR, Irimia M, Karapiperis C, Scouras ZG, Blencowe BJ, Promponas VJ, Ouzounis CA.
Sci Rep. 2014;4:4655.
Biological information extraction and co-occurrence analysis.
Pavlopoulos GA, Promponas VJ, Ouzounis CA, Iliopoulos I.
Methods Mol Biol. 2014;1159:77-92.
Development of an ELISA for the quantification of the C-terminal decapeptide prothymosin α(100-109) in sera of mice infected with bacteria.
Samara P, Kalbacher H, Ioannou K, Radu DL, Livaniou E, Promponas VJ, Voelter W, Tsitsilonis O.
J Immunol Methods. 2013 Sep 30;395(1-2):54-62.
Meeting report: the seventh conference of the hellenic society for computational biology and bioinformatics.
Hamodrakas SJ, Iliopoulos I, Ouzounis CA, Bagos PG, Promponas VJ.
Comput Struct Biotechnol J. 2013;6:e201303006.
GPU technology as a platform for accelerating local complexity analysis of protein sequences.
Papadopoulos A, Kirmitzoglou I, Promponas VJ, Theocharides T.
Annu Int Conf IEEE Eng Med Biol Soc. 2013;2013:2684-7
The chlamydiales pangenome revisited: structural stability and functional coherence.
Psomopoulos FE, Siarkou VI, Papanikolaou N, Iliopoulos I, Tsaftaris AS, Promponas VJ, Ouzounis CA.
Genes (Basel). 2012;3(2):291-319.
A comparative study on filtering protein secondary structure prediction.
Kountouris P, Agathocleous M, Promponas VJ, Christodoulou G, Hadjicostas S, Vassiliades V, Christodoulou C.
IEEE/ACM Trans Comput Biol Bioinform. 2012;9(3):731-9.
LaTcOm: a web server for visualizing rare codon clusters in coding sequences.
Theodosiou A, Promponas VJ.
Bioinformatics. 2012;28(4):591-2.
BioTextQuest: a web-based biomedical text mining suite for concept discovery.
Papanikolaou N, Pafilis E, Nikolaou S, Ouzounis CA, Iliopoulos I, Promponas VJ.
Bioinformatics. 2011;27(23):3327-8.
Spectral density ratio based clustering methods for the binary segmentation of protein sequences: a comparative study.
Ioannou A, Fokianos K, Promponas VJ.
Biosystems. 2010;100(2):132-43.
Gene socialization: gene order, GC content and gene silencing in Salmonella.
Papanikolaou N, Trachana K, Theodosiou T, Promponas VJ, Iliopoulos I.
BMC Genomics. 2009;10:597.
Establishment of a European Network for Urine and Kidney Proteomics.
Vlahou A, Schanstra J, Frokiaer J, El Nahas M, Spasovski G, Mischak H, Domon B, Allmaier G, Bongcam-Rudloff E, Attwood T; European Kidney and Urine Proteomics Network.
J Proteomics. 2008 Oct 7;71(4):490-2.
PRED-GPCR: GPCR recognition and family classification server.
Papasaikas PK, Bagos PG, Litou ZI, Promponas VJ, Hamodrakas SJ.
Nucleic Acids Res. 2004;32(Web Server issue):W380-2.
GeneViTo: visualizing gene-product functional and structural features in genomic datasets.
Vernikos GS, Gkogkas CG, Promponas VJ, Hamodrakas SJ.
BMC Bioinformatics. 2003;4:53.
Evaluation of annotation strategies using an entire genome sequence.
Iliopoulos I, Tsoka S, Andrade MA, Enright AJ, Carroll M, Poullet P, Promponas V, Liakopoulos T, Palaios G, Pasquier C, Hamodrakas S, Tamames J, Yagnik AT, Tramontano A, Devos D, Blaschke C, Valencia A, Brett D, Martin D, Leroy C, Rigoutsos I, Sander C, Ouzounis CA.
Bioinformatics. 2003 Apr 12;19(6):717-26.
PRED-CLASS: cascading neural networks for generalized protein classification and genome-wide applications.
Pasquier C, Promponas VJ, Hamodrakas SJ.
Proteins. 2001;44(3):361-9.
CAST: an iterative algorithm for the complexity analysis of sequence tracts.
Promponas VJ, Enright AJ, Tsoka S, Kreil DP, Leroy C, Hamodrakas S, Sander C, Ouzounis CA.
Bioinformatics. 2000;16(10):915-22.
Reproducibility in genome sequence annotation: the Plasmodium falciparum chromosome 2 case.
Tsoka S, Promponas V, Ouzounis CA.
FEBS Lett. 1999;451(3):354-5.
A novel method for predicting transmembrane segments in proteins based on a statistical analysis of the SwissProt database: the PRED-TMR algorithm.
Pasquier C, Promponas VJ, Palaios GA, Hamodrakas JS, Hamodrakas SJ.
Protein Eng. 1999;12(5):381-5.
CoPreTHi: a Web tool which combines transmembrane protein segment prediction methods.
Promponas VJ, Palaios GA, Pasquier CM, Hamodrakas JS, Hamodrakas SJ.
In Silico Biol. 1999;1(3):159-62.
A web server to locate periodicities in a sequence.
Pasquier CM, Promponas VI, Varvayannis NJ, Hamodrakas SJ.
Bioinformatics. 1998;14(8):749-50.